|
|
Research
1. Lysokine-Mediated Signaling Pathways in Regulating Longevity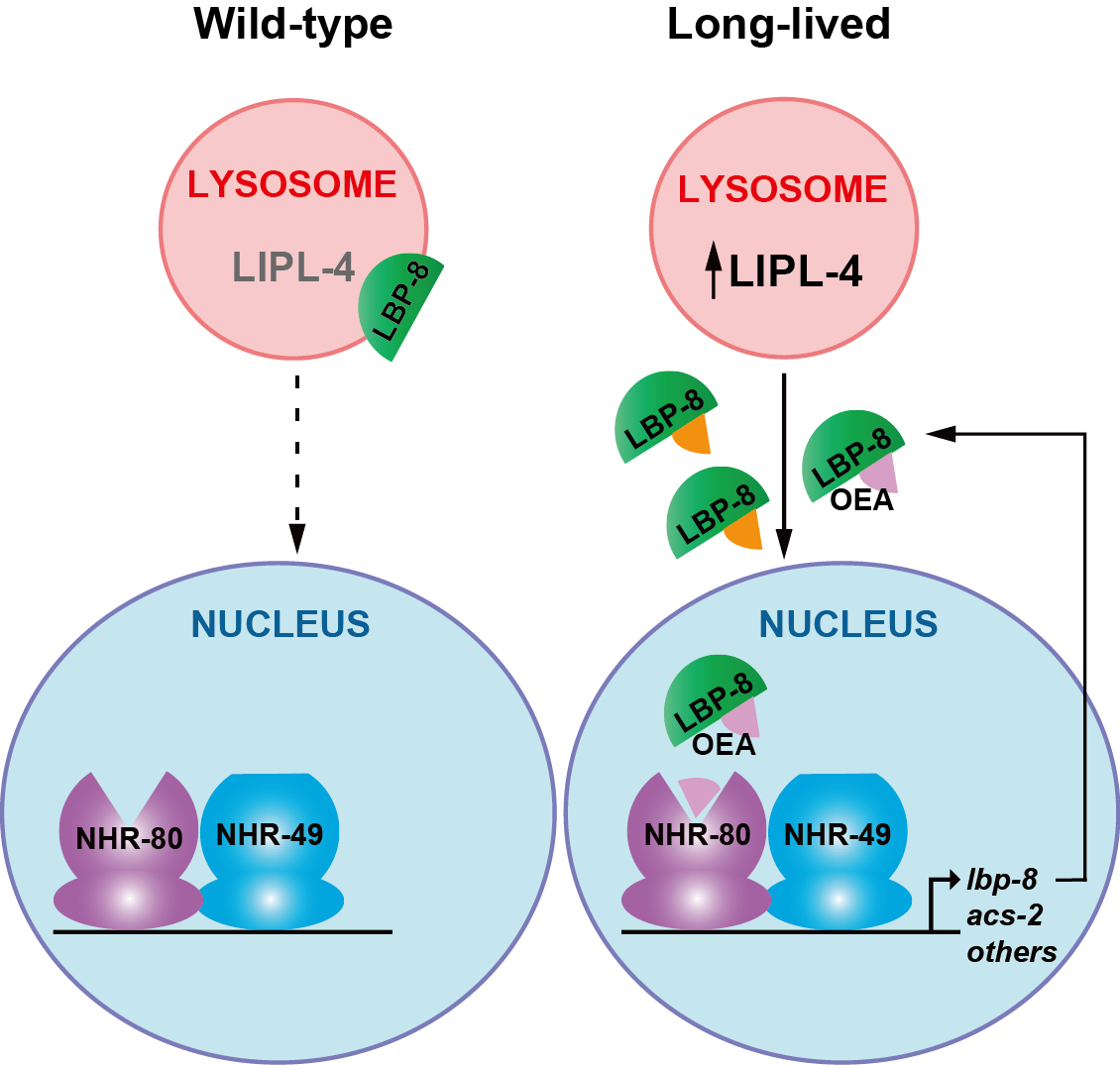
Lysosomes are dynamic organelles for cellular homeostasis, and active sites for metabolite production. Lysosomal hydrolysis generates various metabolites that are delivered to different cellular compartments as structural material and energy fuels. Questioning whether lysosomal hydrolysis also promotes signaling molecules (lysokines) for intra-organellar communication, we made the discovery that in response to lysosomal lipolysis, the lipid chaperone protein LBP-8 translocates from the lysosome to the nucleus, carrying the lipid metabolite oleoylethanolamide to activate nuclear receptors and consequently promote longevity. This previously unknown lysosome-to-nucleus retrograde lipid signaling reveals the tip of the iceberg in lysosome-to-nucleus communication. We are now combining lipodomics, chemical proteomics and functional genomics to systematically discover metabolites and proteins functioning in the lysosome-to-nucleus communication network, to investigate the mechanisms by which metabolites and proteins cooperate, and to decipher the significance of this interorganellar network in regulating cellular homeostasis and organismal healthspan.
2. Signaling Crosstalk between Bacteria and Host Mitochondria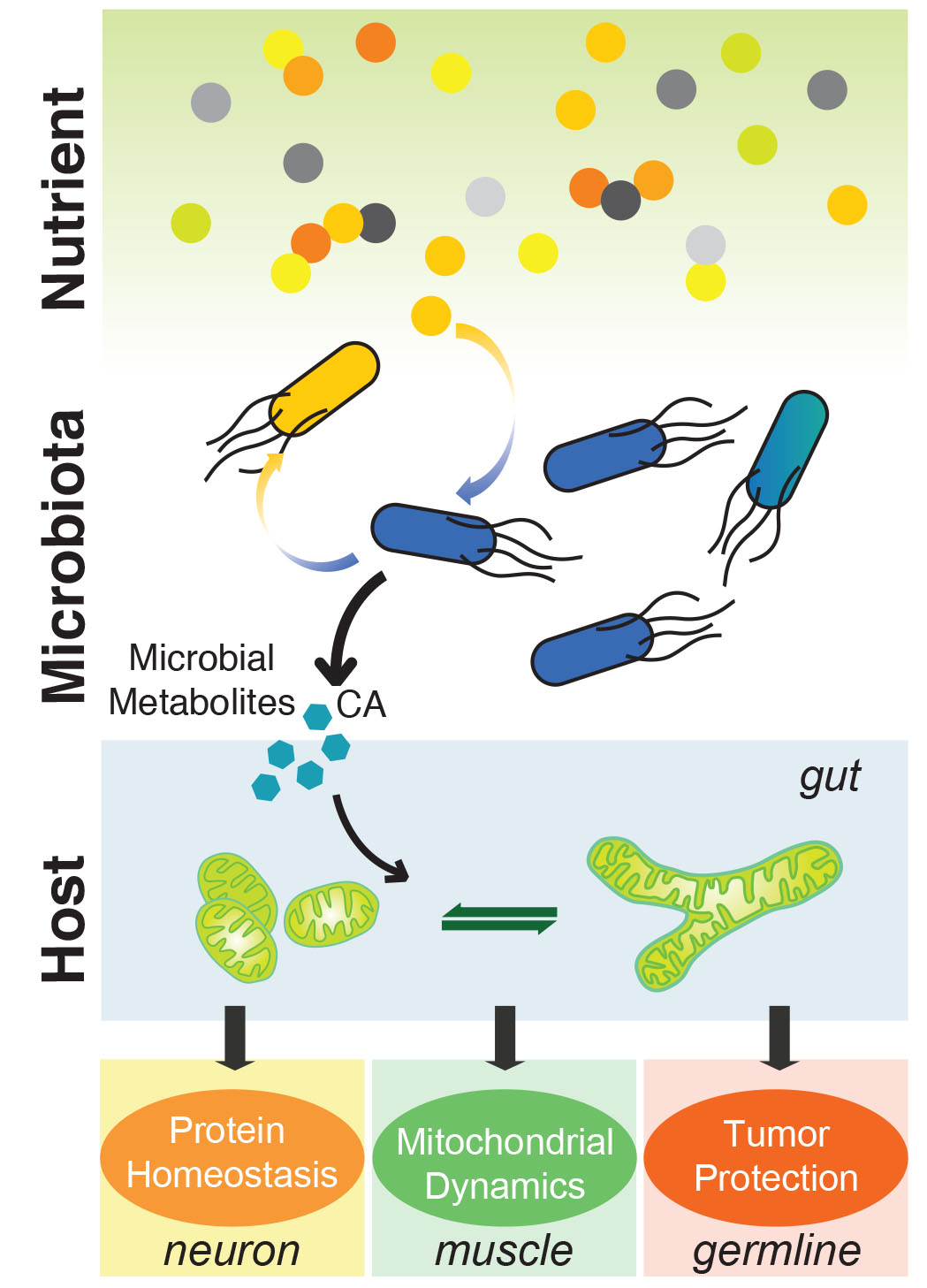
None of the organisms is sterile. Microbes share the same environment with their eukaryotic hosts, and play pivotal roles in modulating the host’s physiological and pathological activities. One of our research areas is to decipher the molecular mechanisms governing the dynamic and constant metabolic communication between microbes and their hosts. We discovered that volatile metabolites derived from bacteria could modify host reproductive strategies via activating a specific neuroendocrine circuit. Our work also revealed that microbe-derived metabolites serve as crucial cues to fine-tune mitochondrial dynamics in the host, and consequently exert an active role in regulating the host’s metabolic fitness and longevity through specific signaling mechanisms. These studies reveal a novel mode of signaling communication between bacteria and mitochondria (the ancient relatives of bacteria in eukaryotic cells). We are now taking the advantage of bacterial genetics and metabolomics and integrating them with C. elegans cellular imaging and functional genomics to systemically investigate this unexplored bacterial-mitochondrial communication system.
3. Cellular and Molecular Mechanisms of Reproductive Aging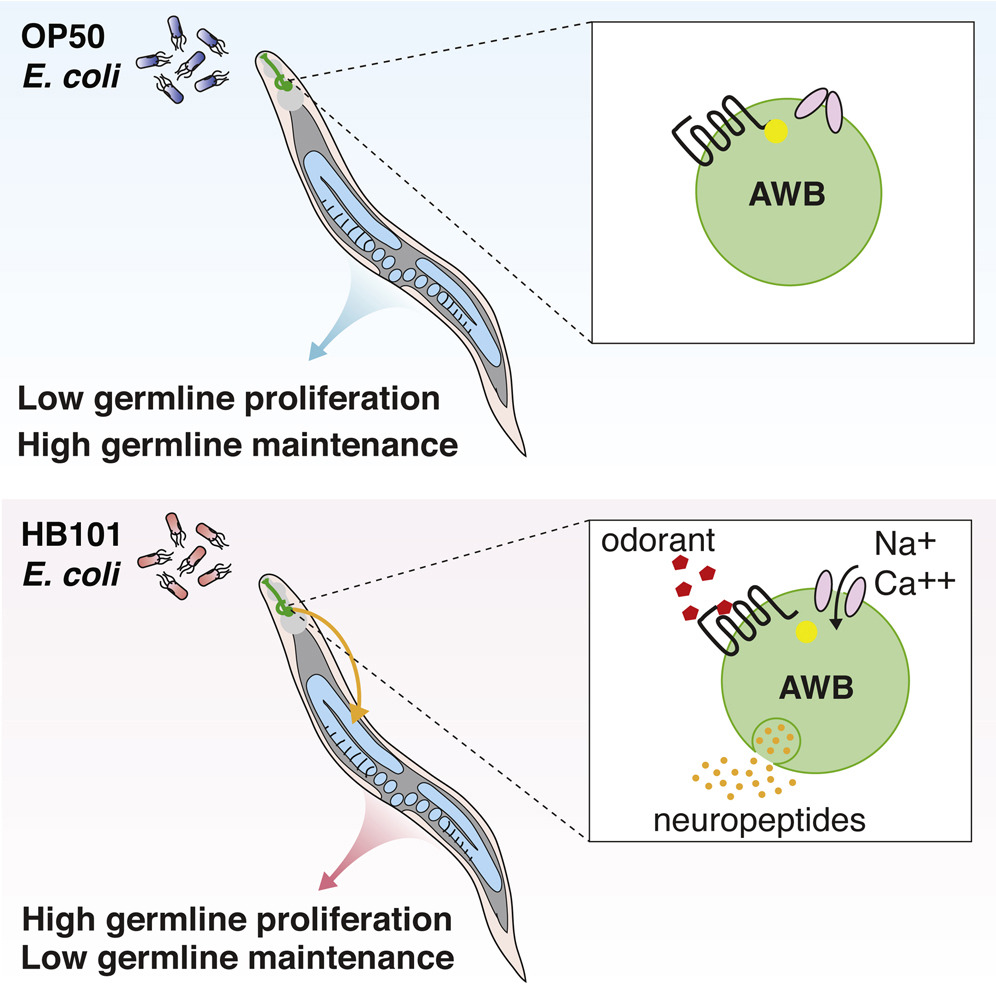
Reproductive health is influenced by both genetic and environmental factors, and declines significantly in women with increasing age. The delay in childbearing is a current trend in western societies, resulting in high risk of subfertility, fetal and perinatal mortality. We aim to reveal both the genetic and environmental regulatory mechanisms of reproductive activities during aging. Through large-scale functional genomic analysis, we identified a series of new genes that regulate reproductive longevity and their crucial interactions with organism longevity. We also discovered a previously unreported neuroendocrine nexus linking olfactory sensation to reproductive longevity. These findings lay the groundwork for further investigation of genetic and environmental impact on reproductive health. We are now taking the advantage of functional genetics, optogenetics and single-cell transcriptome to map the neural circuit in regulating reproductive longevity, decipher neuroendocrine signals linking the nervous and the reproductive systems, and to understand the mechanistic link between reproductive and somatic longevity.
4. New Ways to Study Spatiotemporal Dynamics of Lipid Metabolism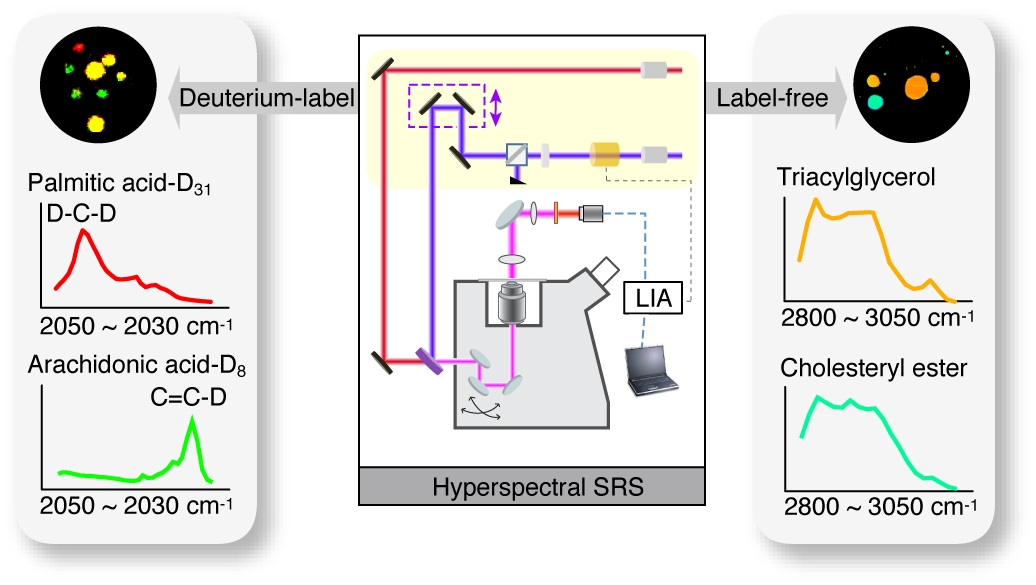
Historically, it has been a technical challenge to directly visualize lipid molecules in vivo. Lipid molecules are intrinsically non-fluorescent, and fluorescence labeling is difficult and often alters their chemical activities. Therefore, their dynamics cannot be accessed by fluorescence microscopy. To tackle this challenge, we have applied stimulated Raman scattering (SRS) microscopy to track spatial-temporal distribution of lipid molecules in living cells and organisms. Using these techniques, we visualized different lipid molecules at the subcellular resolution in real time, revealing their compositional changes in association with various metabolic pathologies. We also demonstrated the power of SRS microscopy systems in functional genomic screens, leading to the discovery of novel lipid regulatory genes. By integrating these chemical imaging platforms with multi-photon fluorescence microscopy systems, we aim to visualize metabolite transportation flux between different cellular compartments and uncover previously unknown interactions between metabolites and proteins. This multiplex imaging platform will provide us unique opportunities to investigate metabolite-directed communication systems as a function of time and space and to characterize the underlying regulatory mechanisms of these spatiotemporal dynamics.
|
